Medical imaging analysis - part 1: A short survey
Some anatomical domains
One can recognize the importance of computer vision in medicine by seeing that there are many anatomical domains using medical images for diagnosis such as brain, skin lesion, blood, lung cancer, cardiac, musculoskeletal, tumor, ...
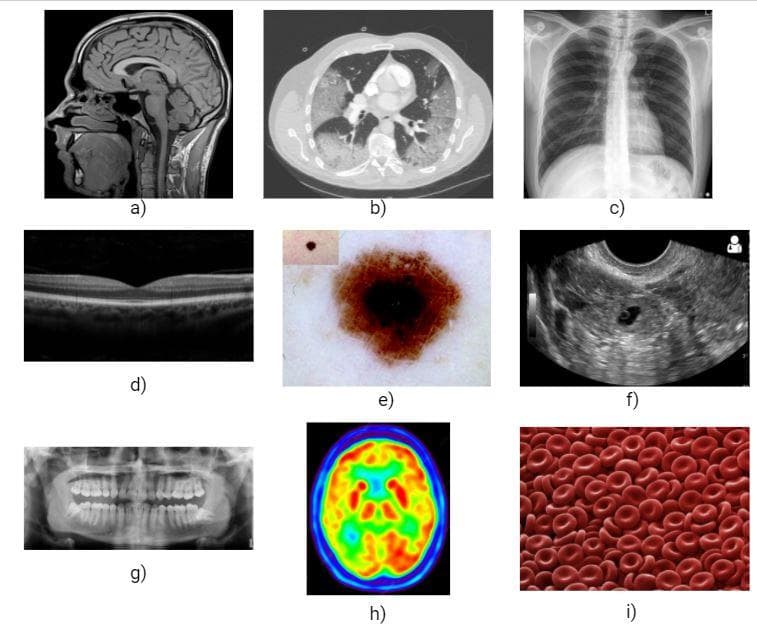
Figure 1: Several types of medical images: a) Brain MRI, b) Body CT, c) Chest X-ray, d) Optical OCT, e) Skin dermoscopy, f) Baby ultrasound, g) Teeth radiograph, h) Brain PET, i) Blood microscopy (Images collected from the Internet).
From figure 1, you can somewhat intuitively grasp the characteristics of several types of medical images. Each type has its own properties and can only be fittingly used in some anatomical domains.
Usually, the diagnosis results from these images of doctors are just correct to some extent. As a result, it will be better if we have computer-aided systems from which supportive predictions can be referred.
Some types of problems
Like in fundamental computer vision, there are also many types of problems in medical imaging [5].
-
Classification:
- Image classification: given an input image, predict whether there is a disease or not.
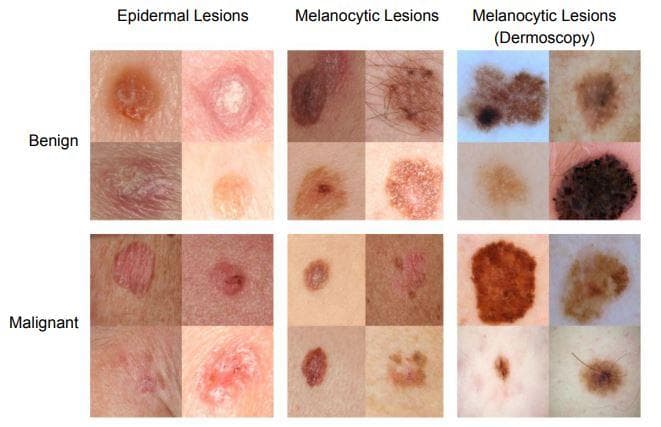
Figure 2: "Skin cancer classification" (Image from [12]).
- Object or lesion classification: instead of classifying on the whole input image, this problem just focuses on a small region/patch of the input image. The categories can be about shape, type, ...
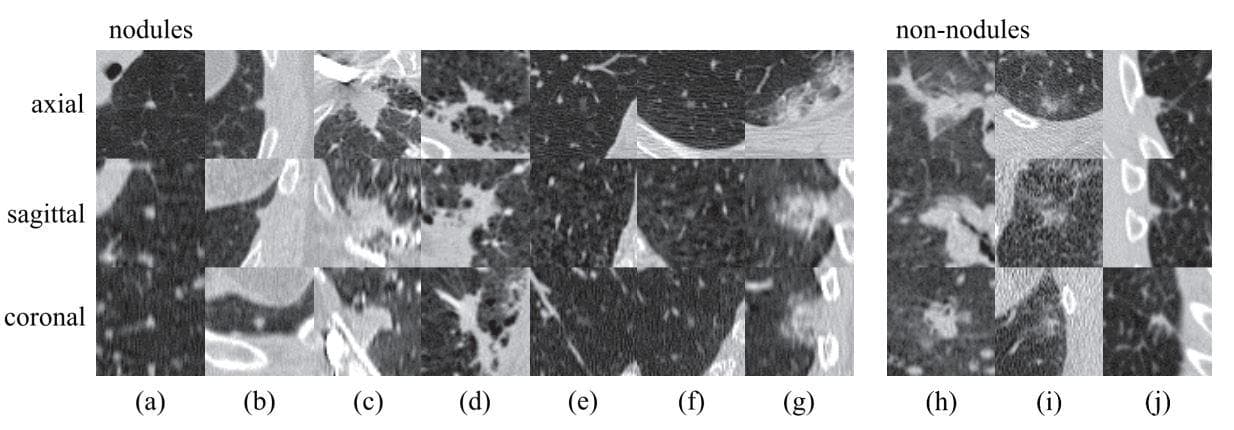
Figure 3: "Regions of pulmonary. Examples of lesions (nodules and non-nodules) in axial, sagittal, and coronal views. Lesions are located in the center of the box (50 50 mm). The left set of images are nodules with a wide range of morphological characteristics: (a)solid nodule, (b) pleural nodule, (c)–(d) large nodules with irregular shape, (e)–(f) subsolid nodules. The right set of images are irregular lesions that are not related to nodules or cancers. These examples illustrate that designing features for accurate detection and classification of nodules may not be trivial" (Image from [13]).
-
Detection - the 2 main types are:
- Organ, region, and landmark localization: this step has usually been done before carrying out the segmentation. One special point of localization in medical imaging is that it is applied a lot to the 3D domain.
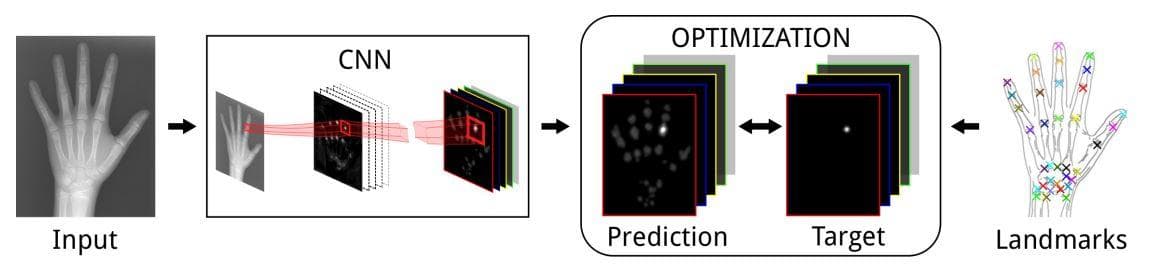
Figure 4: "Landmark localization. Multiple landmark localization by regressing heatmaps in a CNN framework" (Image from [14]).
- Object or lesion detection: This type of task is still a localization but with one additional step that is identification/classification. The difference with image classification is that object classification is processed at the pixel level. Every pixel is classified, so there is a bias towards the non-object classes.
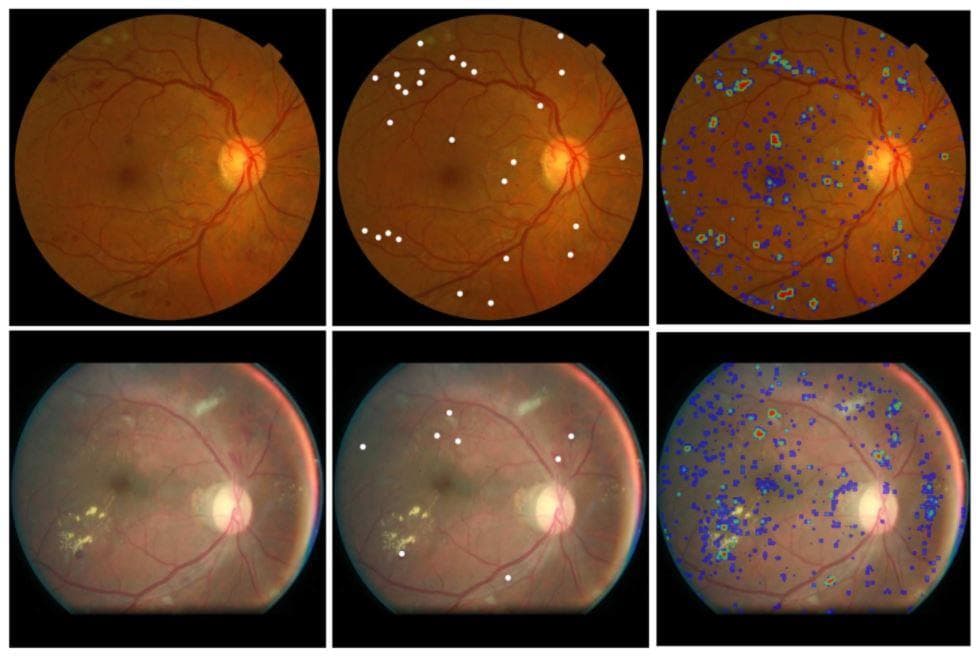
Figure 5: "Left column: example color fundus images from the Kaggle test set. Middle column: reference hemorrhage center locations. Right column: output of the SeS CNN 60" (Image from [15]).
-
Segmentation - the 2 main types are:
- Organ and substructure segmentation: segmenting the object(s) of interest from the medical image is a significant first step in pipelines, which allows us quantitatively analyze the volumes and shapes of discriminating regions.
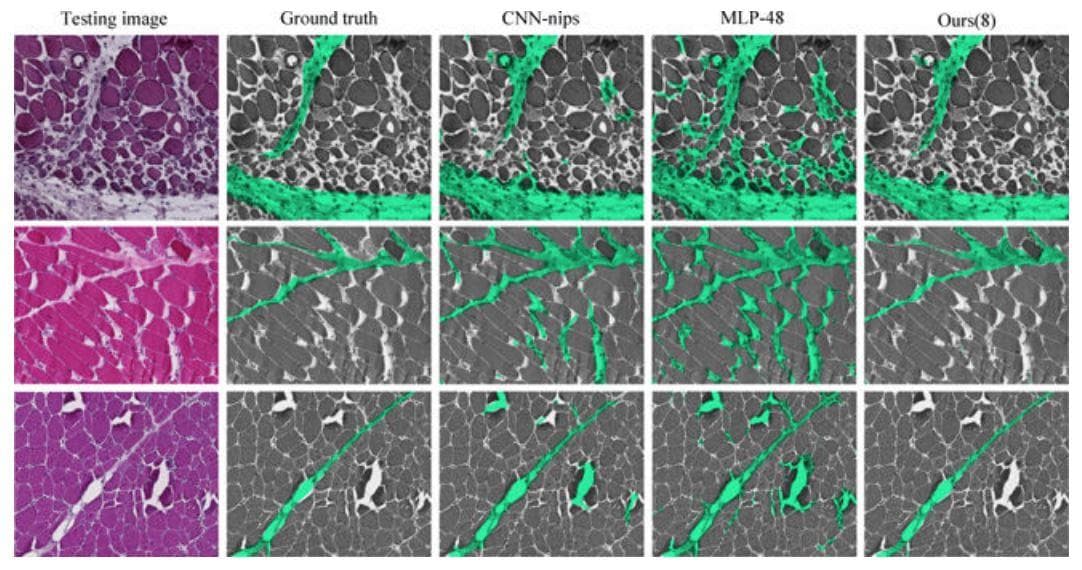
Figure 6: "Perymisum segmentation results on three challenging skeleton muscle images which show strong global structure and demonstrates a lot of appearance similarity between perimysium (true positive) and endo/epimysium (false positive). Comparing with other methods, our results show much better global consistency because it can capture global spatial configurations" (Image from [16]).
- Lesion segmentation: both object detection and substructure segmentation need to be concerned to tackle this problem.
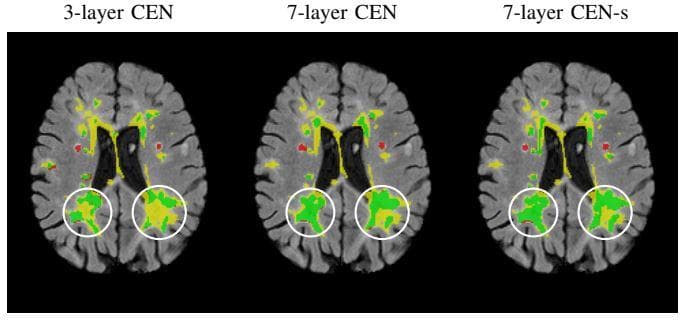
Figure 7: "Detect the white lesions of brain. Yellow, green, and red are the lesion regions. Impact of increasing the depth of the network on the segmentation performance of very large lesions. The true positive, false negative and false positive voxels are highlighted in green, yellow, and red, respectively. The 7- layer CEN, with and without shortcut, is able to segment large lesions much better than the 3-layer CEN due to the increased size of the receptive field. This figure is best viewed in color" (Image from \\[17]).
- Registration (or spatial alignment): is the task of coordinate transformation, which is calculated from the variation of two medical images. According to Wikipedia [18], it “is the process of transforming different sets of data into one coordinate system”. This type of problem is thought by researchers to be more difficult than classification and segmentation.
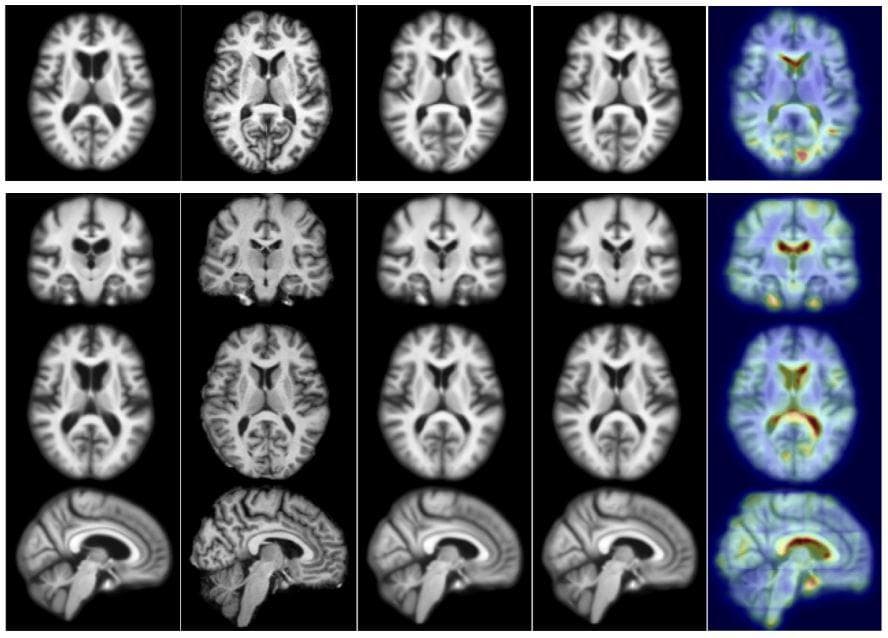
Figure 8: "Test example for 2D (top) and 3D (bottom). From left to right: moving (atlas) image, target image, deformation result by optimizing LDDMM energy, deformation result using 50 samples from the probabilistic network with a stride of 14 and patch pruning, and uncertainty as the square root of the sum of the variances of deformation in all directions mapped on the predicted registration result. The colors indicate the amount of uncertainty (red = high uncertainty, blue = low uncertainty). Best viewed in color" (Image from [19]).
- Image generation/Image enhancement: this type of task can be used for removing blocking elements from images, improving the quality of image, discovering patterns, ...
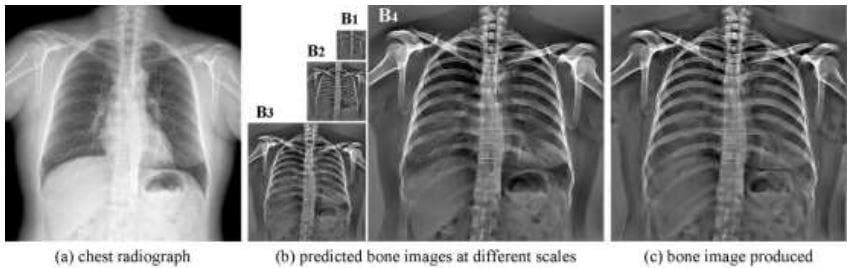
Figure 9: "Example of predicted bone images. (a) Chest radiograph. (b) Bone images at different scales predicted by a 4-level CamsNet. (c) Bone image produced by fusing the predicted multi-scale bone gradients from the CamsNet" (Image from [20]).
By listing some types of problems above, it cannot be denied that the impact of computer vision on medical imaging analysis is very massive.
Handcraft features
Beside Deep Learning, handcraft features like Local Binary Pattern (LBP) [3] or edge-based features [4] have also been used to extract salient information from medical images. Both of the papers are published in 2016.
- “Medical Image Classification via SVM using LBP Features from Saliency-Based Folded Data” [3]: this paper has an image preprocessing stage of using saliency map and image folding to concentrate on the relevant region without losing much surrounding information. From the preprocessed image, LBP features are extracted and then fed into SVM for classification.
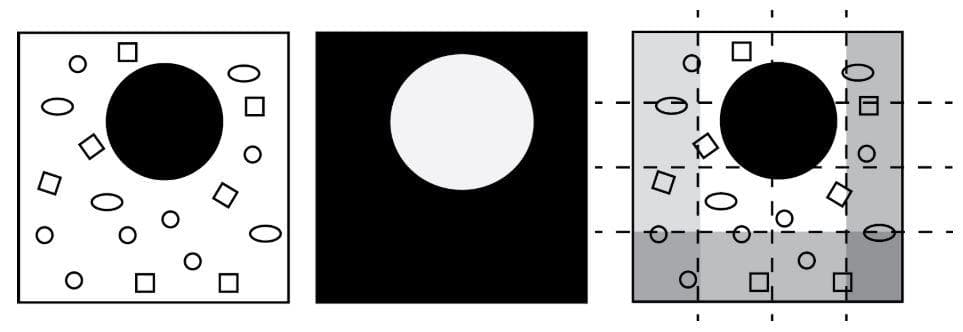
Figure 10: "Schematic illustration of saliency maps and image folding: The input image (left image) is processed to find a salient region (middle image). Subsequently, non-salient regions (right image, gray stripes) are marked to be folded inwardly" (Image from [3]).
- “Classification of medical images using edge-based features and sparse representation” [4]: this paper makes a classification on an 18-category dataset. Each category contains radiological images of a human body part, e.g. skull, hand, breast, pelvis, elbow, etc. Canny edge detection is used on each image, then it will be split into patches. Next, each patch is divided into concentric circular regions. Finally, the mean and variance of pixel values in each circular region are used to build the feature vector. For classification, they make use of the Online Dictionary Learning algorithm [21] to construct a dictionary for each class.
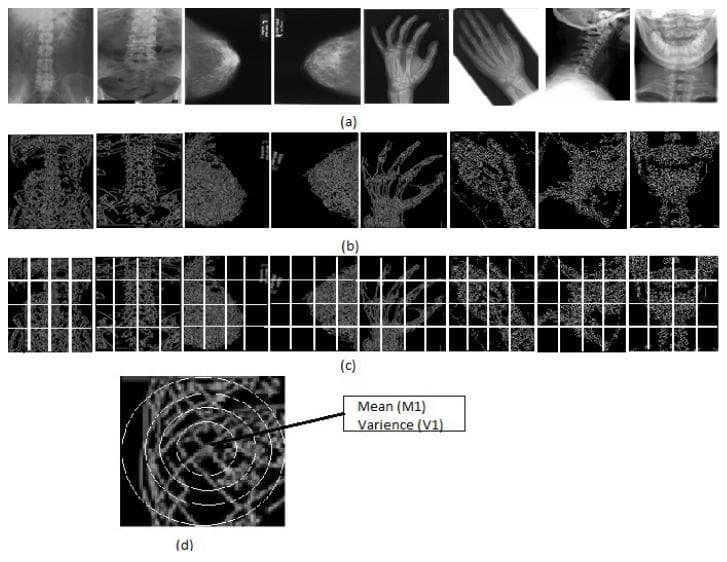
Figure 11: "Includes (a) samples of the IRMA medical images. (b) Coressponding edge images (c) Images are divided into equal size of patches.(d) A patch is divided into concentric circular regions" (Image from [4]).
One can strongly argue for the reason handcrafted features are still used in 2016 (after the explosion of Deep Learning) by the assumption that medical image contains low-level information (like the shape of a small region of the skin or the appearance of some small details, ...) which is good for discriminating between different categories and therefore the local property of handcrafted features still plays an essential role. In addition to that, handcraft methods are usually very lightweight; therefore, they can pass the time requirements of real-world applications.
Survey of Deep Learning models [5]
Supervised models: CNN and RNN
Convolutional neural network (CNN) is usually used the most in medical imaging analysis. Next is Recurrent neural network (RNN), Long short term memory (LSTM), or Gated recurrent unit (GRU) though they were initially proposed for one-dimensional sequence input. For medical image segmentation, the 2 convolutional network architectures mentioned in [5] are fully convolutional neural network (fCNN) and U-net.
Unsupervised models: AE, DBN, and RBM
These methods learn to extract features by reconstructing the input image without the need for labels. Conventionally, the middle latent embedding is seen as containing salient information for classification.
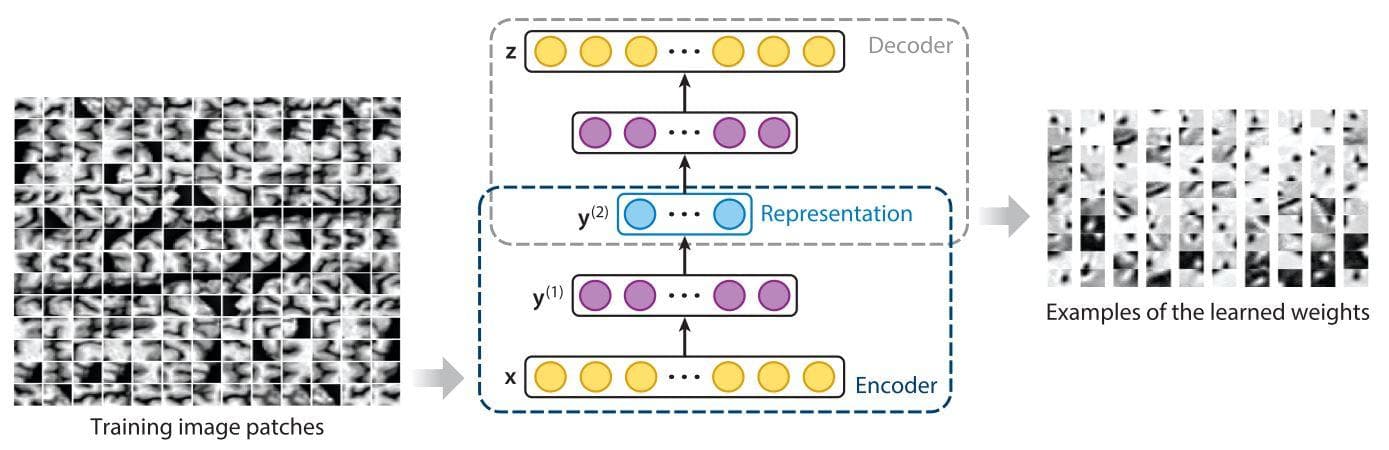
Figure 12: "Construction of a deep encoder–decoder via a stacked auto-encoder and visualization of the learned feature representations. The blue circles represent high-level feature representations. The yellow and purple circles indicate the correspondence between layers in the encoder and decoder" (Image from [11]).
Besides CNN and RNN, the 3 next artificial neural networks that are mostly used for feature extraction from medical images are Autoencoder (AE), Deep Belief Network (DBN), and Restricted Boltzmann Machine (RBM). All of these 3 networks are in the category of unsupervised learning and have the purpose of learning meaningful representations/features from unlabelled data.
To see the properties, pros, and cons of each of the 3 networks, you should read from the table below.
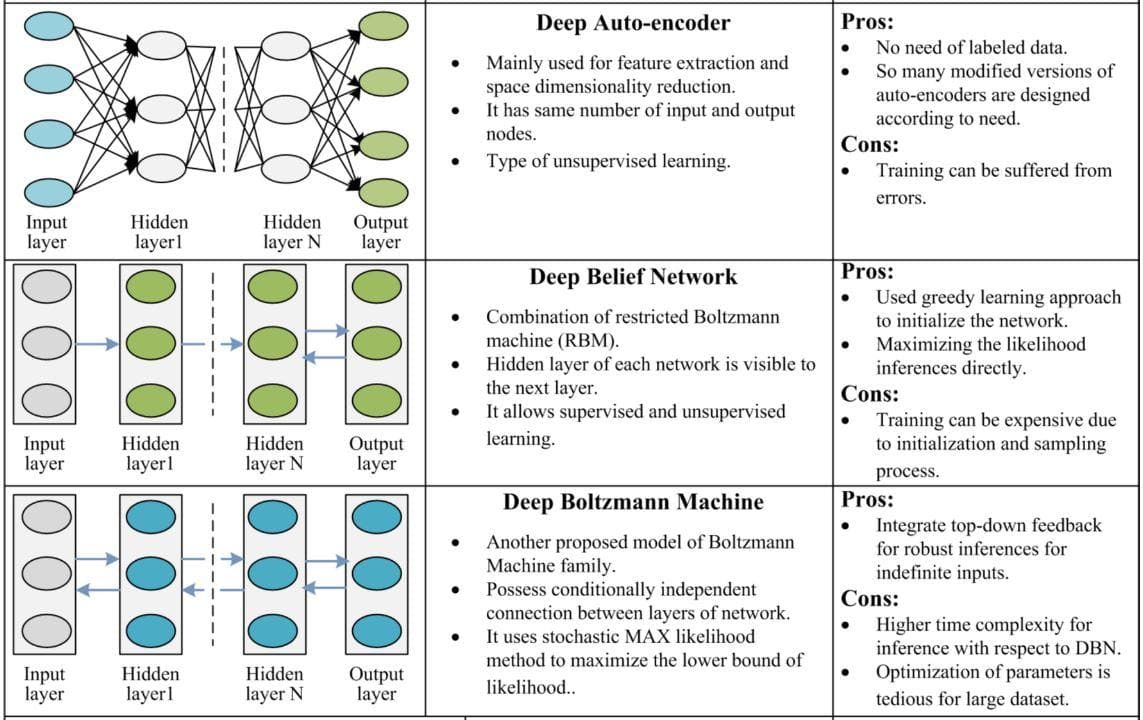
Figure 13: Comparison of Autoencoder, Deep Belief Network and Restricted Boltzman Machine (Image from [10]).
The labels may be considered as a bias source, which prevents the true learning of the true information from input data. With the 3 methods above, we don’t need labels because the labels are the input itself. They try to generate the latent embedding from the input image and reconstruct the image again from that latent embedding. This latent embedding, by chance, has the possibility to become a high-quality feature/representation.
Deep learning - Convolutional neural network [1]
One of the earliest papers that uses deep learning for medical imaging is [1]. This paper uses a convolutional neural network (CNN) with a shallow convolution layer for medical image patch classification for interstitial lung disease (ILD). The authors suppose that extracting feature descriptors by CNN is more generalizable, automatic, and efficient than previous feature descriptors.
The public ILD dataset is used for evaluation. Its sample images have patches of 5 classes/categories like in the Figure below.
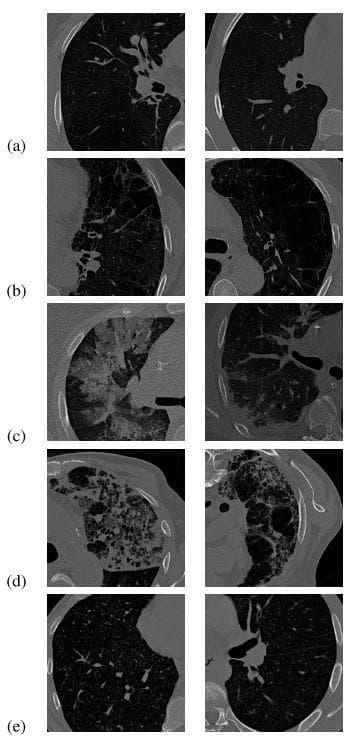
Figure 14: "Two example HRCT images are shown for each category of ILD tissues: (a) normal; (b) emphysema; (c) ground glass; (d) fibrosis; (e) micronodules (Image from [1])."
The used network architecture of this paper is very simple, which just includes 1 convolutional layer, 1 max-pooling layer, and 3 fully connected layers.
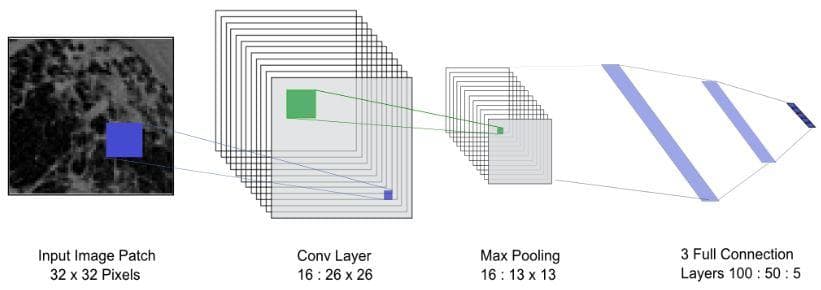
Figure 15: The used network architecture (Image from [1]).
They compare the network performances with 3 other feature extraction methods: SIFT, rotation-invariant local binary pattern (LBP) with 3 resolutions, and Restricted Boltzmann Machine (RBM). Then, the extracted features are brought to SVM for classification. The classification results show that CNN is better than the others in both precision and recall.
Some newer potential approaches
In addition to CNN, there have been some recent approaches that try to improve the performance of medical imaging analysis in all kinds of aspects.
A generative adversarial net [22] which includes encoder network and decoder networks has also been used for medical imaging analysis [7]. One can immediately think of the use of it for image data synthesis because of the lack of medical data for training or for reconstruction and denoising. However, GAN is also a strong candidate for feature extraction. It can also be used for classification or segmentation. See the figure below for some applications of GAN in medical.
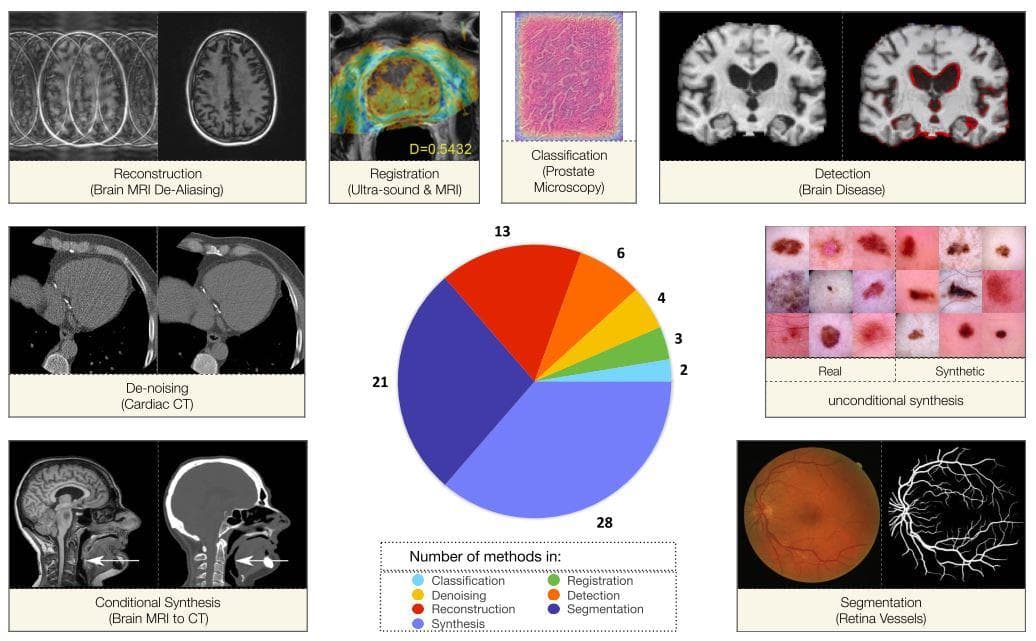
Figure 16: "The pie chart of distribution of papers and visual examples of GAN functionality among the different applications." The GAN papers are chosen by the authors (Image from [7]).
The lack of labeled data definitely triggers the use of semi-supervised methods. [8] proposes a relation-driven semi-supervised framework to deal with the problem of the small number of labeled medical data in image classification. To tackle the problem of insufficiency in high-quality expertise-required dense segmentation labels, [23] presents a teacher-student framework for organ and lesion segmentation with the combination of both dense segmentation labels and bounding box labels.
Explainable AI [6]
Medical imaging analysis is a multi-disciplinary domain that requires the collaborations of both AI researchers and doctors. In general, for people in different domains or especially industry product or business aims, it is always vital to explain the prediction results of a model to related parties. If we cannot explain why a model gives a result, then the reliability of that model can not be verified. This requirement strictly relates to the morality problem.
There are several taxonomies that have been proposed to discriminate different explainability methods. One of them, which is from [6], is shown in the figure below
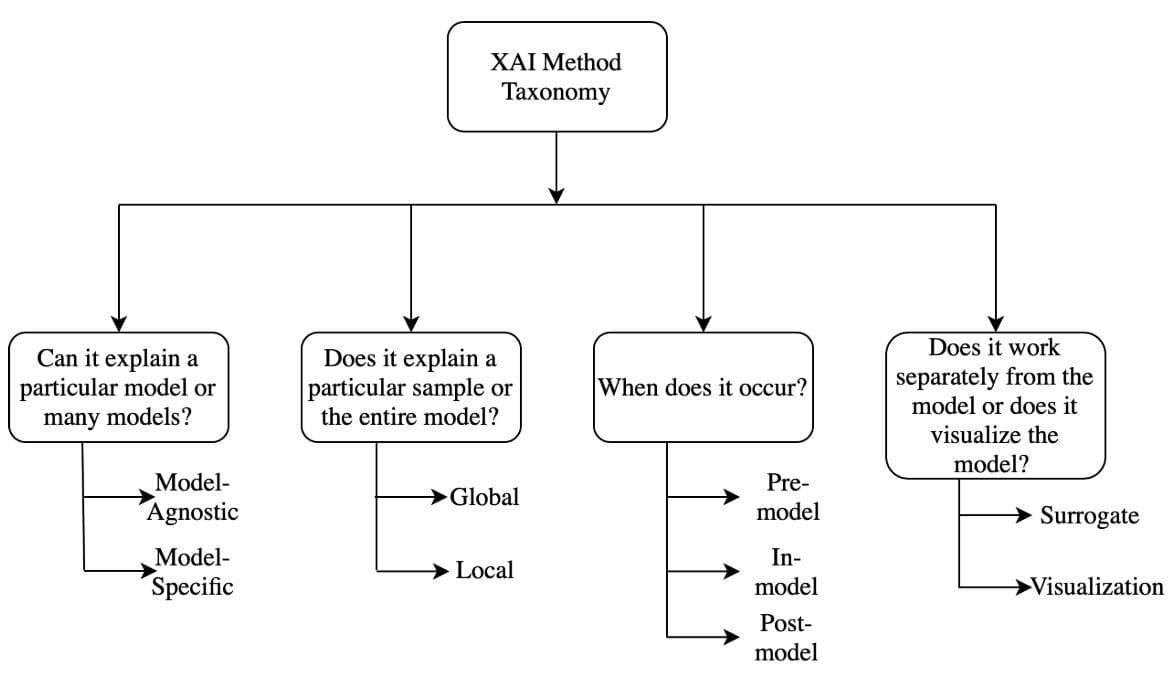
Figure 17: "Taxonomy of XAI methods" (Image from [6]).
Most visualization methods are constructed on the basis of highlighting what pixels, regions of input influence the final prediction result. From this type of knowledge, a researcher can check if the deep model has been learned in the right way to give the final result and find better solutions if it does not work correctly.
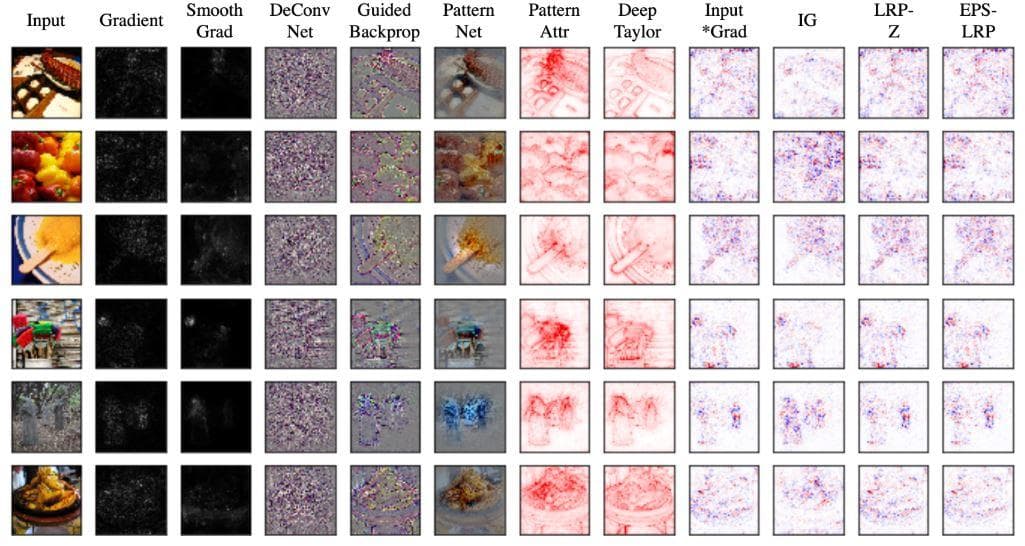
Figure 18: Some visualization methods (Image from [6]).
Achieving effective explainability AI methods has a very significant effect on the progress of medical imaging analysis. It will leverage all other medical researches and especially allow doctors to get insights from and know what a neural network does to conclude the final prediction result.
Medical image dataset [2]
Dataset is another key problem in medical imaging analysis. There have been some papers specifically published to bring up this difficulty alone. For instance, the paper titled "How much data is needed to train a medical image deep learning system to achieve necessary high accuracy?" [2] presents research on how to choose the optimum size of a dataset that is required to get good classification results.
According to [9], it categorizes the dataset problem into two smaller problems for the segmentation task:
- Scarce annotations: the lack of expertise-required annotations.
- Weak annotations: the lack of high-quality dense annotations.
- The lack of medical images: this is an additional problem added by us. Indeed, it can be seen as the most important problem. Because if you don't even have data, you can do nothing with deep learning.
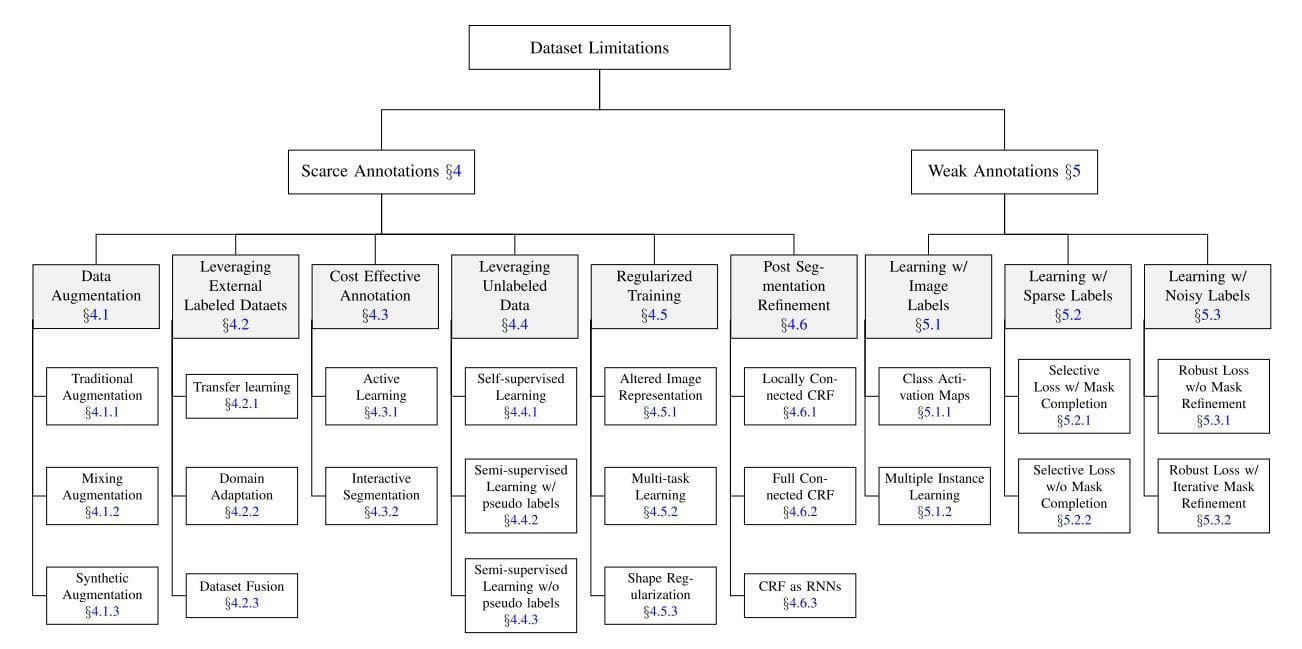
Figure 19: Taxonomy of dataset limitations: scarce annotations and weak annotations. The strategies (gray boxes) and their particular instances to deal with each problem are also provided. (Image from [9]).
As you can see from the figure above, the most traditional way to deal with the lack of annotations or training samples is data augmentation. Next, come transfer learning and domain adaptation to take advantage of other available labeled datasets for learning. Some approaches that use unlabeled data like self-supervised learning or semi-supervised learning are noticed as well. For weak annotations, all three strategies are about learning labels: learning image labels, learning sparse labels and learning noisy labels.
What does weak annotation mean in the segmentation context? Occasionally, dense annotation is used where each pixel is annotated with respect to the category it belongs to. That perfect annotation method is very expensive. Weak annotation tries to relax the labor work of dense annotation. The figure below shows some annotation methods for the three types of learning labels of weak annotation.
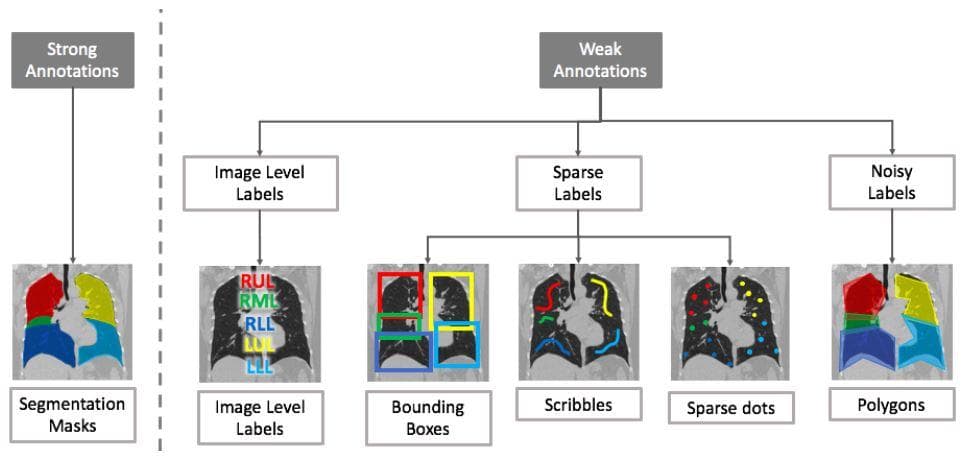
Figure 20: . (Image from [9]).
The weak annotations are clearly easier to have than the strong annotations.
Conclusion
Data is very important in deep learning. In medical imaging, the importance of data is even greater than that. Due to some privacy and right ownership, medical data cannot be accessed by a large number of people when compared to the general open dataset. Besides, a good feature extraction method is also very crucial, but not sufficient; it also needs to be explainable for reliability and morality. Currently, there have been many signs of progress in all of the three mentioned problems and soon the future of medicine will definitely inherit their advantages to make an extent to people’s health movement.
References
[1] Li, Q., Cai, W., Wang, X., Zhou, Y., Feng, D. D., & Chen, M. (2014). Medical image classification with convolutional neural network. 2014 13th International Conference on Control Automation Robotics and Vision, ICARCV 2014, 844–848. https://doi.org/10.1109/ICARCV.2014.7064414
[2] Cho, J., Lee, K., Shin, E., Choy, G., & Do, S. (2015). How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? http://arxiv.org/abs/1511.06348
[3] Camlica, Z., Tizhoosh, H. R., & Khalvati, F. (2016). Medical image classification via SVM using LBP features from saliency-based folded data. Proceedings - 2015 IEEE 14th International Conference on Machine Learning and Applications, ICMLA 2015, 128–132. https://doi.org/10.1109/ICMLA.2015.131
[4] Srinivas, M., & Mohan, C. K. (2016). Classification of medical images using edge-based features and sparse representation. ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing - Proceedings, 2016-May(March), 912–916. https://doi.org/10.1109/ICASSP.2016.7471808
[5] Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., van der Laak, J. A. W. M., van Ginneken, B., & Sánchez, C. I. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42(1995), 60–88. https://doi.org/10.1016/j.media.2017.07.005
[6] Singh, A., Sengupta, S., & Lakshminarayanan, V. (2020). Explainable deep learning models in medical image analysis. Journal of Imaging, 6(6), 1–19. https://doi.org/10.3390/JIMAGING6060052
[7] Kazeminia, S., Baur, C., Kuijper, A., van Ginneken, B., Navab, N., Albarqouni, S., & Mukhopadhyay, A. (2020). GANs for medical image analysis. Artificial Intelligence in Medicine, 109, 1–40. https://doi.org/10.1016/j.artmed.2020.101938
[8] Liu, Q., Yu, L., Luo, L., Dou, Q., & Heng, P. A. (2020). Semi-Supervised Medical Image Classification With Relation-Driven Self-Ensembling Model. IEEE Transactions on Medical Imaging, 39(11), 3429–3440. https://doi.org/10.1109/TMI.2020.2995518
[9] Tajbakhsh, N., Jeyaseelan, L., Li, Q., Chiang, J. N., Wu, Z., & Ding, X. (2020). Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Medical Image Analysis, 63(2019). https://doi.org/10.1016/j.media.2020.101693
[10] Bhatt, C., Kumar, I., Vijayakumar, V., Singh, K. U., & Kumar, A. (2021). The state of the art of deep learning models in medical science and their challenges. Multimedia Systems, 27(4), 599–613. https://doi.org/10.1007/s00530-020-00694-1
[11] Zhang, Y., Gorriz, J. M., & Dong, Z. (2021). Deep learning in medical image analysis. Journal of Imaging, 7(4), NA. https://doi.org/10.3390/jimaging7040074
[12] Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., & Thrun, S. (2017). Dermatologist-level classification of skin cancer with deep neural networks. nature, 542(7639), 115-118.
[13] Setio, A. A. A., Ciompi, F., Litjens, G., Gerke, P., Jacobs, C., Van Riel, S. J., ... & Van Ginneken, B. (2016). Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE transactions on medical imaging, 35(5), 1160-1169.
[14] Payer, C., Štern, D., Bischof, H., & Urschler, M. (2016, October). Regressing heatmaps for multiple landmark localization using CNNs. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 230-238). Springer, Cham.
[15] Van Grinsven, M. J., van Ginneken, B., Hoyng, C. B., Theelen, T., & Sánchez, C. I. (2016). Fast convolutional neural network training using selective data sampling: Application to hemorrhage detection in color fundus images. IEEE transactions on medical imaging, 35(5), 1273-1284.
[16] Xie, Y., Zhang, Z., Sapkota, M., & Yang, L. (2016, October). Spatial clockwork recurrent neural network for muscle perimysium segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 185-193). Springer, Cham.
[17] Brosch, T., Tang, L. Y., Yoo, Y., Li, D. K., Traboulsee, A., & Tam, R. (2016). Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE transactions on medical imaging, 35(5), 1229-1239.
[18][image registration](https://en.wikipedia.org/wiki/Image_registration), Wikipedia.
[19] Yang, X., Kwitt, R., & Niethammer, M. (2016). Fast predictive image registration. In Deep Learning and Data Labeling for Medical Applications (pp. 48-57). Springer, Cham.
[20] Yang, W., Chen, Y., Liu, Y., Zhong, L., Qin, G., Lu, Z., ... & Chen, W. (2017). Cascade of multi-scale convolutional neural networks for bone suppression of chest radiographs in gradient domain. Medical image analysis, 35, 421-433.
[21] Mairal, J., Bach, F., Ponce, J., & Sapiro, G. (2009, June). Online dictionary learning for sparse coding. In Proceedings of the 26th annual international conference on machine learning (pp. 689-696).
[22] Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., ... & Bengio, Y. (2014). Generative adversarial nets. Advances in neural information processing systems, 27.
[23] Sun, L., Wu, J., Ding, X., Huang, Y., Wang, G., & Yu, Y. (2020). A Teacher-Student Framework for Semi-supervised Medical Image Segmentation From Mixed Supervision. arXiv preprint arXiv:2010.12219.